What Technology is used in Comparing DNA?
Techy Mind
---
The Technology used in comparing DNA is called DNA Sequencing. Sequencing is the process of determining the order of the nucleotides in a stretch of DNA. The most common type of sequencing is known as Sanger sequencing, which uses a technique called chain termination.
In this method, DNA polymerase creates a new strand of DNA while adding dideoxynucleotides (ddNTPs) to the mix. These ddNTPs lack one hydrogen bond that their natural counterparts possess, meaning they can only be added to the end of a growing strand of DNA. When one is incorporated into the new strand, it causes the DNA polymerase to fall off, terminating further growth.
 |
| Technology is used in Comparing DNA |
DNA comparison is a process of identifying the similarities and differences between two strands of DNA. This can be done by sequencing the DNA and then aligning the sequences to look for areas of similarity and difference. Several different algorithms can be used to compare DNA sequences, and new methods are being developed all the time.
What Technology is Used in DNA Sequencing?
DNA sequencing is the process of determining the order of the four chemical blocks (nucleotides) in a strand of DNA. It involves any method or technology that can read the sequence of these four nucleotide bases -cytosine, thymine, gaunine, and adenine. Rapid DNA sequencing methods have greatly accelerated biological and medical researches and discoveries.
The first DNA sequence was determined in 1977 by Frederick Sanger using what was then called the chain-termination method. In this technique, enzymes are used to create fragments of varying lengths from a template strand of DNA. These fragments are then separated by electrophoresis, and their sizes are determined by comparison to known standards.
The sequence of the template strand can be inferred from these size differences. While Sanger’s technique was revolutionary at the time, it was very laborious and could only be used to sequence relatively short stretches of DNA. The development of automated sequencers in the 1980s made it possible to rapidly sequence much longer stretches of DNA.
These instruments use fluorescence to detect the incorporation of labelled nucleotides into growing strands of DNA during replication. By reading the sequence generated by each machine cycle, researchers can determine the order of nucleotides in a sample of DNA.
What Technique is Used to Analyze Genomes?
DNA sequencing is the process of determining the order of the chemical bases (nucleotides) in a DNA molecule. Sequencing is used to identify mutations, track the spread of diseases, and determine an organism’s evolutionary history. There are several methods for DNA sequencing, but the most common is called Sanger sequencing.
In Sanger sequencing, DNA is copied in a test tube using enzymes. The copy contains radioactive nucleotides, which are then separated by size using gel electrophoresis. A machine then reads the sequence of nucleotides from the gel.
Sanger sequencing can produce long sequences, but it is expensive and time-consuming. Newer methods, such as next-generation sequencing (NGS), can generate millions of sequences at once. NGS is used to sequence genomes, which are all the DNA in an organism.
Genome sequencing has led to important discoveries about many species, including humans.
What is Sequencing Technology?
Sequencing technology is used to determine the order of nucleotides in a DNA or RNA molecule. This information can be used to identify genes, mutations and other features of the genome. There are several different sequencing technologies available, each with its benefits and drawbacks.
The most common sequencing technologies are Sanger sequencing, next-generation sequencing (NGS) and whole-genome sequencing (WGS). Sanger sequencing is the traditional method of DNA sequencing. It is highly accurate but also very expensive and time-consuming.
NGS is a newer technology that can sequence large amounts of DNA much faster than Sanger sequencing. However, it is less accurate than Sanger sequencing and has a higher error rate. WGS is the newest type of DNA sequencing technology.
It sequences an entire genome at once and can provide detailed information about all the genes in a person’s DNA. However, it is also very expensive and time-consuming.
Which Sequencing Technology is the Best?
There are many different sequencing technologies available, and it can be hard to know which one is the best for your needs. Here is a brief overview of some of the most popular sequencing technologies to help you make a decision:
1. Sanger Sequencing: Sanger sequencing is the traditional method of DNA sequencing. It involves using chemicals to break down DNA into smaller pieces, which are then sequenced by a machine. This technology is very accurate and can sequence long stretches of DNA. However, it is also quite slow and expensive.
2. Next Generation Sequencing (NGS): NGS is a newer type of sequencing that can generate large amounts of data very quickly and cheaply. However, it is not as accurate as Sanger sequencing and cannot sequence very long stretches of DNA.
3. Single-Molecule Real-Time (SMRT) Sequencing: SMRT sequencing is a newer technology that can sequence extremely long stretches of DNA accurately without the need for chemicals or machines. However, it is much more expensive than other methods.
4. Nanopore Sequencing: Nanopore sequencing is a newer technology that sequences DNA by passing it through tiny pores in a membrane.
DNA Sequencing Technologies
DNA sequencing technologies have revolutionized the field of biology. By allowing us to read the sequence of nucleotides in DNA, these technologies have given us unprecedented insight into the genetic code. There are a variety of different DNA sequencing technologies available, each with its advantages and disadvantages.
The most common DNA sequencing technology is Sanger sequencing, also known as chain termination sequencing. This enzyme causes the DNA replication process to terminate randomly, producing fragments of varying lengths.
These fragments are then separated by size using gel electrophoresis and sequenced using traditional methods. Sanger sequencing is relatively fast and inexpensive, but it has several drawbacks. First, it can only generate short sequences (usually less than 1000 bases).
Second, it requires a large amount of starting material (several micrograms). Finally, it is not suitable for sequences with repeats or other complex features. Next-generation sequencing (NGS) technologies address some of the limitations of Sanger sequencing.
NGS platforms can generate millions or even billions of reads in a single run, making them well suited for genome-scale projects. In addition, NGS platforms can produce longer reads (up to several thousand bases), which help assemble genomes from scratch or for characterizing long repeats and other complex features. Finally, many NGS platforms do not require as much starting material as Sanger sequencing; some can work with as little as a few nanograms of DNA.
Next-Generation Sequencing
The race to sequence the human genome was one of the most important scientific achievements of the 20th century. The Human Genome Project, which began in 1990, took more than a decade and $3 billion to complete. But that was just the beginning.
In the years since, sequencing technology has advanced by leaps and bounds, becoming faster, cheaper, and more accurate. Today, it’s possible to sequence an entire human genome in a matter of days for less than $1,000. This rapid progress has sparked a new era of genomic medicine, in which DNA sequencing is being used to diagnose and treat diseases.
Next-generation sequencing (NGS) is providing insights into the causes of rare genetic disorders, cancer, and other complex diseases. It’s also being used to match patients with clinical trials for precision medicines or find suitable donors for bone marrow transplants. To learn more about next-generation sequencing and its implications for health care, check out this infographic from Harvard Medical School.
Genomic Techniques
The Human Genome Project, completed in 2003, was a monumental achievement in genomic science. This international research effort was the first to sequence the entire human genome, providing scientists with a detailed map of our DNA. Today, thanks to advances in technology, we can do much more than just sequence genomes.
We can now use a variety of techniques to study genes and their functions at a level of detail that was unimaginable just a few years ago. Here are some of the most popular genomic techniques currently being used by researchers:
1. next-generation sequencing (NGS): This technique is used to quickly sequence large stretches of DNA or even an entire genome. NGS is often used in cancer research to identify mutations that may be driving tumour growth.
2. Gene Expression Analysis: By measuring the activity levels of individual genes, this technique can give us insight into which genes are turned on or off in different tissues or under different conditions. Gene expression analysis is often used to study how diseases like cancer develop and progress.
3. Epigenetics: Epigenetics refers to changes in gene activity that are not caused by changes in the DNA sequence itself. These changes can be passed down from one generation to the next and play an important role in the development and disease. Epigenetic studies are helping us better understand how environmental factors can influence our health.
4) Single-Cell Genomics: This relatively new field of research uses special techniques to analyze the genomes of individual cells rather than populations of cells.
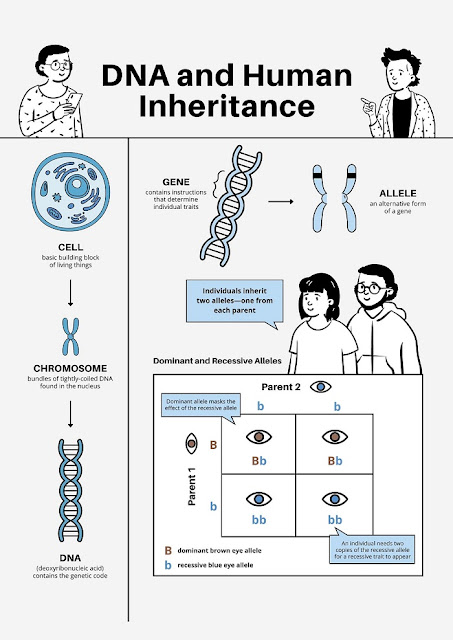 |
| DNA and Human Inheritance |
Importance of DNA Sequencing
The Human Genome Project was an international effort to sequence the entire human genome, which was completed in 2003. The resulting data has provided researchers with a powerful tool to identify genes associated with disease and to develop new diagnostic and therapeutic approaches. The sequencing of other genomes, including those of model organisms such as mice and fruit flies, has also yielded important insights into biology.
For example, comparisons of the mouse and human genomes have helped to identify genes involved in cancer. Such work would not have been possible without advances in DNA sequencing technology. DNA sequencing is now used routinely in many areas of biomedical research, from studying the genetics of cancer to tracing the origins of infectious diseases.
It is also being used increasingly in clinical settings, for example, to screen newborn babies for genetic disorders or to match patients with treatments that target their specific genetic makeup.
Illumina Sequencing
As the sequencing of genomes becomes more routine, there is an ever-growing demand for higher throughput and lower costs. This has spurred the development of new sequencing technologies, one of which is Illumina sequencing. Illumina sequencing is a next-generation sequencing technology that can sequence an entire human genome in just a few days.
The key advantage of Illumina sequencing over other technologies is its high throughput – it can sequence up to 1 billion bases per run. This makes it ideal for large-scale projects such as genome-wide association studies (GWAS). The other key advantage of Illumina sequencing is its low cost.
Unlike traditional Sanger sequencing, which can cost upwards of $100,000 per genome, Illumina sequencing can be done for less than $5,000 per genome. This makes it affordable for research projects that require the sequences of many genomes. While Illumina sequencers are not yet available for purchase by individual researchers, they are widely used by commercial providers such as 23andMe and AncestryDNA.
These companies use Illumina sequencers to provide DNA testing services to consumers.
Functional Genomics Techniques
Functional genomics is the study of the function of all genes in a genome. It can be used to determine which genes are responsible for specific functions, and how they work together. Functional genomics techniques include:
1. DNA microarrays: DNA microarrays can be used to measure the expression of thousands of genes at once. This allows for the identification of genes that are differentially expressed in different conditions or tissues.
2. RNA interference (RNAi): RNAi is a technique that can be used to shut down specific genes. This can be used to determine the function of those genes.
3. CRISPR-Cas9: CRISPR-Cas9 is a tool that can be used to edit genomes. This allows for the study of gene function in a way that was not possible before.
Conclusion
The technology used in comparing DNA is called DNA sequencing, gel electrophoresis. This process separates the DNA by size and then uses a special dye to make it visible. The smaller the piece of DNA, the faster it will move through the gel.
Post a Comment
Post a Comment